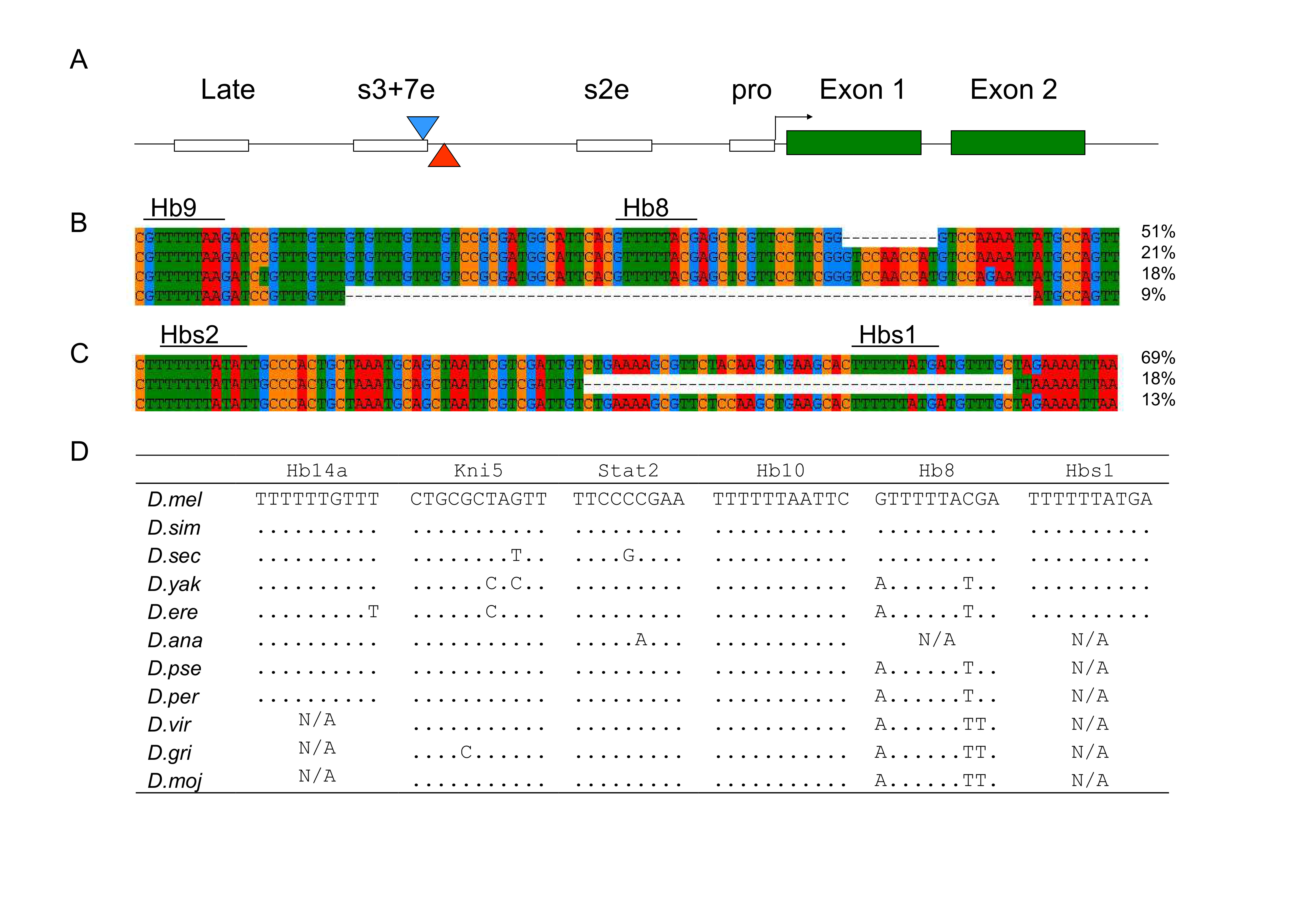
Naturally occurring deletions of Hunchback binding sites in the even-skipped stripe 3+7 enhancer
Our manuscript on deletion variations in regulatory elements was published in Plos One.
Naturally occurring deletions of Hunchback binding sites in the even-skipped stripe 3+7 enhancer
Arnar Palsson, Natalia Wesolowska, Sigrún Reynisdóttir, Michael Z. Ludwig and Martin Kreitman
Faculty of Life and Environmental Sciences, University of Iceland, Reykjavik, Iceland.
Institute of Biology, University of Iceland, Reykjavik, Iceland
Department of Ecology and Evolution, University of Chicago, Chicago, IL, USA.
Changes in regulatory DNA contribute to phenotypic differences within and between taxa. Comparative studies show that many transcription factor binding sites (TFBS) are conserved between species and functional studies reveal that some mutations segregating within species alter TFBS function. Single base and insertions/deletion polymorphism are rare in characterized regulatory elements in Drosophila melanogaster. Experimentally defined TFBS are nearly devoid of segregating mutations and evolutionarily conserved. For instance, 8 of 11 Hunchback binding sites in the stripe 3+7 enhancer of even-skipped are conserved between D. melanogaster and Drosophila virilis. Oddly, we found a 72 bp deletion that removes one of those binding sites (Hb8), segregating within D. melanogaster. Furthermore, a 45 bp deletion polymorphism in the spacer between the stripe 3+7 and stripe 2 enhancers, removes another predicted Hunchback site. These two deletions are separated by ~250 bp, sit on distinct haplotypes, and segregate at appreciable frequency. The Hb8del is at 5 to 35% frequency in the new world, but also has cosmopolitan distribution. There is depletion of sequence variation on the Hb8del carrying haplotype. Quantitative genetic tests indicate that Hb8del affects developmental time, but not viability of offspring. The Eve expression pattern differs between inbred lines, but the stripe 3 and 7 boundaries are seemingly not affected by the Hb8del. The deletions of those two Hb binding sites in the eve regulatory elements do not appear to be deleterious to D. melanogaster. The data reveal segregating variation for a characterized TFBS, which may reflect evolutionary turnover due to drift or co-evolution.
Figure 1 of the manuscript illustrates the main findings.