Now published in Molecular Ecology
Whole genome sequencing reveals how plasticity and genetic differentiation underlie sympatric morphs of Arctic charr.
https://onlinelibrary.wiley.com/doi/10.1111/mec.70085
ABSTRACT
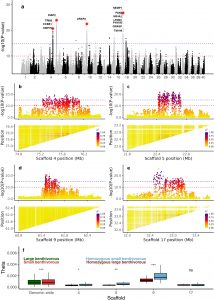
Salmonids have a remarkable ability to form sympatric morphs after postglacial colonisation of freshwater lakes. These morphs often differ in morphology, feeding and spawning behaviour. Here, we explored the genetic basis of morph differentiation in Arctic charr (n = 283) by first establishing a high-quality reference genome and then using this in whole genome sequencing of distinct morphs present in two Norwegian and two Icelandic lakes. The four lakes represent the spectrum of genetic differentiation between morphs from one lake with no genetic differentiation between morphs, implying phenotypic plasticity, to two lakes with locus-specific genetic differentiation, implying incomplete reproductive isolation, and one lake with strong genome-wide divergence consistent with complete reproductive isolation. As many as 12 putative inversions ranging from 0.45 to 3.25 Mbp in size segregated among the four morphs present in one lake, Thingvallavatn, and these contributed significantly to the genetic differentiation among morphs. None of the putative inversions were found in any of the other lakes, but there were cases of partial haplotype sharing in similar morph contrasts in other lakes. Our findings are consistent with a highly polygenic basis of morph differentiation with population-specific selection on alleles linked to the development of similar morph phenotypes. The results support a model where morph differentiation is first established through phenotypic plasticity, leading to niche expansion and separation. This may be followed by gradual development of reproductive isolation, locus-specific differentiation and eventually complete reproductive isolation and genome-wide divergence.